TASBE Calibrated Flow Cytometry¶
This module combines all of the other calibrated flow cytometry modules (autofluorescence, bleedthrough compensation, bead calibration, and channel translation) into one easy-use-interface.
- Channels
Which channels are you calibrating?
- Autofluorescence
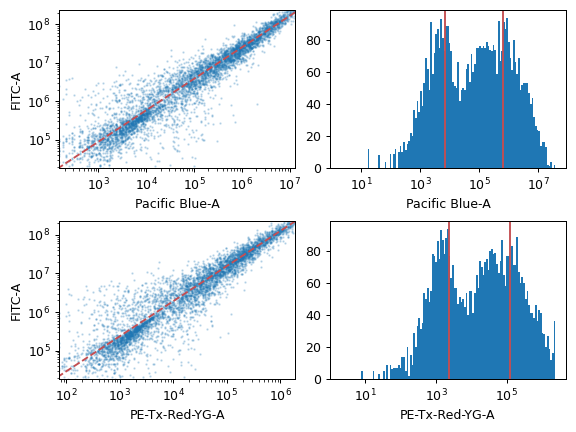
- Blank File
The FCS file with the blank (unstained or untransformed) cells, for autofluorescence correction.
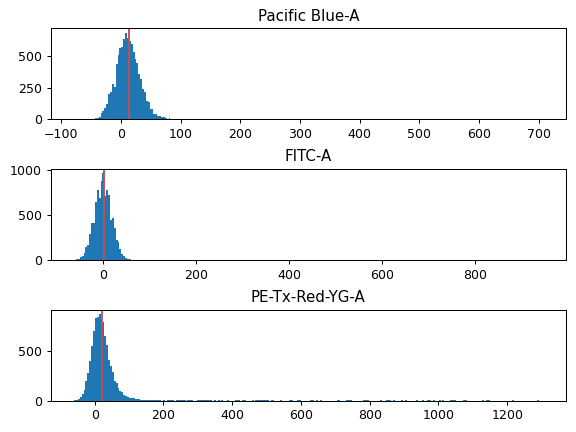
- Bleedthrough Correction
A list of single-color controls to use in bleedthrough compensation. There’s one entry per channel to compensate.
- Channel
The channel that this file is the single-color control for.
- File
The FCS file containing the single-color control data.
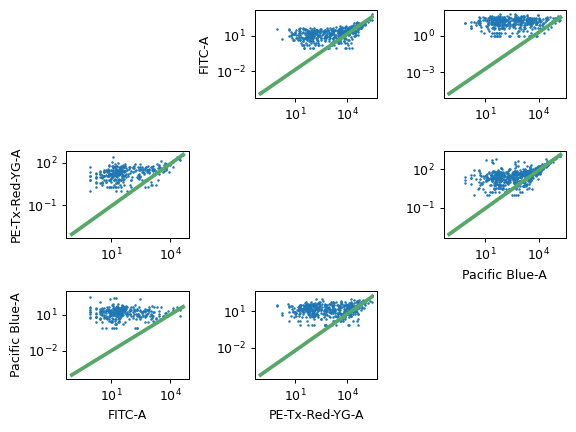
- Bead Calibration
The beads that you used for calibration. Make sure to check the lot number as well!
The FCS file containing the bead data.
The unit (such as MEFL) to calibrate to.
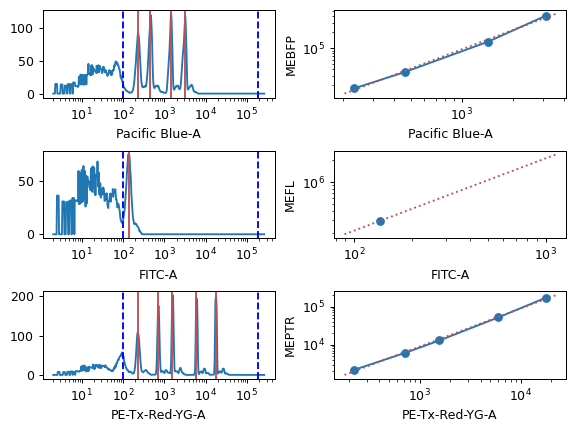
- Peak Quantile
The minimum quantile required to call a peak in the bead data. Check the diagnostic plot: if you have peaks that aren’t getting called, decrease this. If you have “noise” peaks that are getting called incorrectly, increase this.
- Peak Threshold
The minumum brightness where the module will call a peak.
- Peak Cutoff
The maximum brightness where the module will call a peak. Use this to remove peaks that are saturating the detector.
- Color Translation
- To Channel
Which channel should we rescale all the other channels to?
- Use mixture model?
If this is set, the module will try to separate the data using a mixture-of-Gaussians, then only compute the translation using the higher population. This is the kind of behavior that you see in a transient transfection in mammalian cells, for example.
- Translation list
Each pair of channels must have a multi-color control from which to compute the scaling factor.