cytoflow.operations.color_translation¶
The color_translation module has two classes:
ColorTranslationOp – translates the measurements in one channel to
the same scale as another channel, using a two- or three-color control to
estimate the transfer function.
ColorTranslationDiagnostic – a diagnostic view showing how the
ColorTranslationOp operation estimated its parameters.
- class cytoflow.operations.color_translation.ColorTranslationOp[source]¶
Bases:
traits.has_traits.HasStrictTraitsTranslate measurements from one color’s scale to another, using a two-color or three-color control.
To use, set up the
controlsdictionary with the channels to convert and the FCS files to compute the mapping. Callestimateto parameterize the module; check that the plots look good by calling theColorTranslationDiagnostic.plotmethod of theColorTranslationDiagnosticinstance returned bydefault_view; then callapplyto apply the translation to anExperiment.- controls¶
Two-color controls used to determine the mapping. They keys are tuples of from-channel and to-channel. The values are FCS files containing two-color constitutive fluorescent expression data for the mapping.
- Type
Dict((Str, Str), File)
- mixture_model¶
If
True, try to model the from channel as a mixture of expressing cells and non-expressing cells (as you would get with a transient transfection), then weight the regression by the probability that the the cell is from the top (transfected) distribution. Make sure you check the diagnostic plots to see that this worked!- Type
Bool (default = False)
- linear_model¶
Set this to
Trueto get a scaling that is strictly multiplicative, mirroring the TASBE approach. Do check the diagnostic plot, though, to see how well (or poorly) your model fits the data.- Type
Bool (default = False)
- control_conditions¶
Occasionally, you’ll need to specify the experimental conditions that the bleedthrough tubes were collected under (to apply the operations in the history.) Specify them here. The key is a tuple of channel names; the value is a dictionary of the conditions (same as you would specify for a
cytoflow.operations.import_op.Tube)- Type
Dict((Str, Str), Dict(Str, Any))
Notes
In the TASBE workflow, this operation happens after the application of
AutofluorescenceOpandBleedthroughLinearOp. The entire operation history of theExperimentthat is passed toestimateis replayed on the control files incontrols, so they are also corrected for autofluorescence and bleedthrough, and have metadata for subsetting.Examples
Create a small experiment:
>>> import cytoflow as flow >>> import_op = flow.ImportOp() >>> import_op.tubes = [flow.Tube(file = "tasbe/mkate.fcs")] >>> ex = import_op.apply()
Create and parameterize the operation
>>> color_op = flow.ColorTranslationOp() >>> color_op.controls = {("Pacific Blue-A", "FITC-A") : "tasbe/rby.fcs", ... ("PE-Tx-Red-YG-A", "FITC-A") : "tasbe/rby.fcs"} >>> color_op.mixture_model = True
Estimate the model parameters
>>> color_op.estimate(ex)
Plot the diagnostic plot
>>> color_op.default_view().plot(ex)
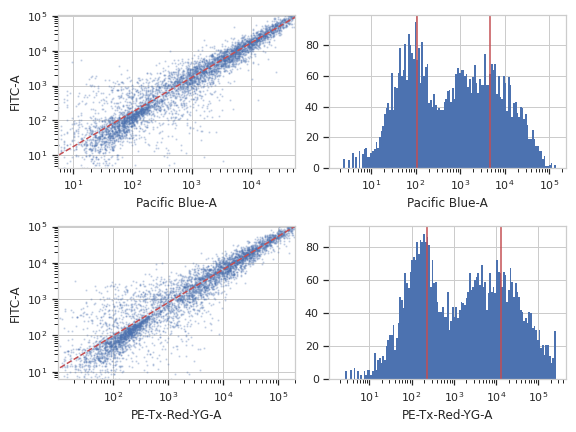
Apply the operation to the experiment
>>> ex = color_op.apply(ex)
- estimate(experiment, subset=None)[source]¶
Estimate the mapping from the two-channel controls
- Parameters
experiment (
Experiment) – TheExperimentused to check the voltages, etc. of the control tubes. Also the source of the operation history that is replayed on the control tubes.subset (Str) – A Python expression used to subset the controls before estimating the color translation parameters.
- apply(experiment)[source]¶
Applies the color translation to an experiment
- Parameters
experiment (
Experiment) – the old_experiment to which this op is applied- Returns
a new experiment with the color translation applied. The corrected channels also have the following new metadata:
channel_translation : Str Which channel was this one translated to?
channel_translation_fn : Callable (
pandas.Series–>pandas.Series) The function that translated this channel- Return type
- default_view(**kwargs)[source]¶
Returns a diagnostic plot to see if the bleedthrough spline estimation is working.
- Returns
A diagnostic view, call
ColorTranslationDiagnostic.plotto see the diagnostic plots- Return type
- class cytoflow.operations.color_translation.ColorTranslationDiagnostic[source]¶
Bases:
traits.has_traits.HasStrictTraits- name¶
The instance name (for serialization, UI etc.)
- Type
Str
- op¶
The op whose parameters we’re viewing
- Type
Instance(
ColorTranslationOp)
- subset¶
A Python expression specifying a subset of the events in the control FCS files to plot
- Type