cytoflow.operations.autofluorescence¶
The cytoflow.operations.autofluorescence module contains two classes:
AutofluorescenceOp – corrects an Experiment for
autofluorescence
AutofluorescenceDiagnosticView – a diagnostic to make sure that
AutofluorescenceOp estimated its parameters correctly.
- class cytoflow.operations.autofluorescence.AutofluorescenceOp[source]¶
Bases:
traits.has_traits.HasStrictTraitsApply autofluorescence correction to a set of fluorescence channels.
The
estimatefunction loads a separate FCS file (not part of the inputExperiment) and computes the untransformed median and standard deviation of the blank cells. Then,applysubtracts the median from the experiment data.To use, set the
blank_fileproperty to point to an FCS file with unstained or nonfluorescing cells in it; set thechannelsproperty to a list of channels to correct.applyalso adds theaf_medianandaf_stdevmetadata to the corrected channels, representing the median and standard deviation of the measured blank distributions.- channels¶
The channels to correct.
- Type
List(Str)
- blank_file¶
The filename of a file with “blank” cells (not fluorescent). Used to
estimatethe autofluorescence.- Type
File
- blank_file_conditions¶
Occasionally, you’ll need to specify the experimental conditions that the blank tube was collected under (to apply the operations in the history.) Specify them here.
- Type
Dict
Examples
Create a small experiment:
>>> import cytoflow as flow >>> import_op = flow.ImportOp() >>> import_op.tubes = [flow.Tube(file = "tasbe/rby.fcs")] >>> ex = import_op.apply()
Create and parameterize the operation
>>> af_op = flow.AutofluorescenceOp() >>> af_op.channels = ["Pacific Blue-A", "FITC-A", "PE-Tx-Red-YG-A"] >>> af_op.blank_file = "tasbe/blank.fcs"
Estimate the model parameters
>>> af_op.estimate(ex)
Plot the diagnostic plot
>>> af_op.default_view().plot(ex)
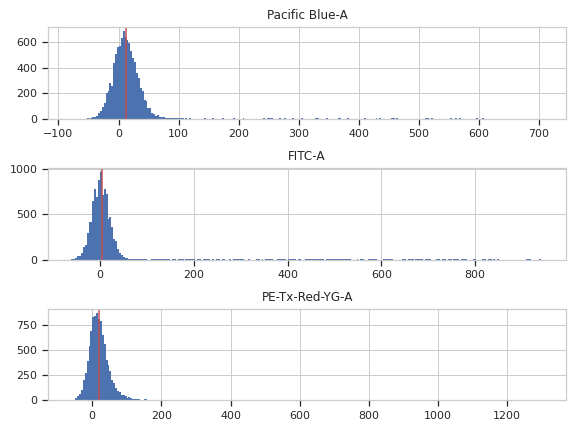
Apply the operation to the experiment
>>> ex2 = af_op.apply(ex)
- estimate(experiment, subset=None)[source]¶
Estimate the autofluorescence from
blank_filein channels specified inchannels.- Parameters
experiment (
Experiment) – The experiment to which this operation is appliedsubset (Str (default = “”)) – An expression that specifies the events used to compute the autofluorescence
- apply(experiment)[source]¶
Applies the autofluorescence correction to channels in an experiment.
- Parameters
experiment (
Experiment) – the experiment to which this op is applied- Returns
a new experiment with the autofluorescence median subtracted. The corrected channels have the following metadata added to them:
af_median : Float The median of the non-fluorescent distribution
af_stdev : Float The standard deviation of the non-fluorescent distribution
- Return type
- default_view(**kwargs)[source]¶
Returns a diagnostic plot to see if the autofluorescence estimation is working.
- Returns
An diagnostic view, call
AutofluorescenceDiagnosticView.plotto see the diagnostic plots- Return type
- class cytoflow.operations.autofluorescence.AutofluorescenceDiagnosticView[source]¶
Bases:
traits.has_traits.HasStrictTraitsPlots a histogram of each channel, and its median in red. Serves as a diagnostic for the autofluorescence correction.
- op¶
The
AutofluorescenceOpwhose parameters we’re viewing. Set automatically if you created the instance usingAutofluorescenceOp.default_view.- Type
Instance(
AutofluorescenceOp)