cytoflow.views.parallel_coords¶
-
class
cytoflow.views.parallel_coords.ParallelCoordinatesView[source]¶ Bases:
cytoflow.views.base_views.BaseNDViewPlots a parallel coordinates plot. PC plots are good for multivariate data; each vertical line represents one attribute, and one set of connected line segments represents one data point.
-
channels¶ The channels to view
Type: List(Str)
-
scale¶ Re-scale the data in the specified channels before plotting. If a channel isn’t specified, assume that the scale is linear.
Type: Dict(Str : {“linear”, “logicle”, “log”})
-
xfacet, yfacet Set to one of the
conditionsin theExperiment, and a new row or column of subplots will be added for every unique value of that condition.Type: String
-
huefacet¶ Set to one of the
conditionsin the in theExperiment, and a new color will be added to the plot for every unique value of that condition.Type: String
Examples
Make a little data set.
>>> import cytoflow as flow >>> import_op = flow.ImportOp() >>> import_op.tubes = [flow.Tube(file = "Plate01/RFP_Well_A3.fcs", ... conditions = {'Dox' : 10.0}), ... flow.Tube(file = "Plate01/CFP_Well_A4.fcs", ... conditions = {'Dox' : 1.0})] >>> import_op.conditions = {'Dox' : 'float'} >>> ex = import_op.apply()
Plot the PC plot.
>>> flow.ParallelCoordinatesView(channels = ['B1-A', 'V2-A', 'Y2-A', 'FSC-A'], ... scale = {'Y2-A' : 'log', ... 'V2-A' : 'log', ... 'B1-A' : 'log', ... 'FSC-A' : 'log'}, ... huefacet = 'Dox').plot(ex)
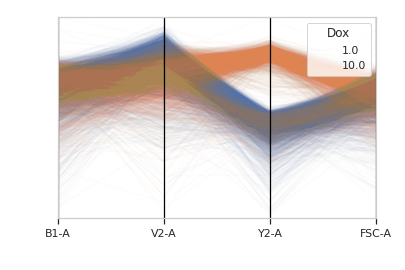
-
plot(experiment, **kwargs)[source]¶ Plot a faceted parallel coordinates plot
Parameters: - experiment (Experiment) – The
Experimentto plot using this view. - title (str) – Set the plot title
- xlabel, ylabel (str) – Set the X and Y axis labels
- huelabel (str) – Set the label for the hue facet (in the legend)
- legend (bool) – Plot a legend for the color or hue facet? Defaults to True.
- sharex, sharey (bool) – If there are multiple subplots, should they share axes? Defaults to True.
- row_order, col_order, hue_order (list) – Override the row/column/hue facet value order with the given list. If a value is not given in the ordering, it is not plotted. Defaults to a “natural ordering” of all the values.
- height (float) – The height of each row in inches. Default = 3.0
- aspect (float) – The aspect ratio of each subplot. Default = 1.5
- col_wrap (int) – If xfacet is set and yfacet is not set, you can “wrap” the subplots around so that they form a multi-row grid by setting col_wrap to the number of columns you want.
- sns_style ({“darkgrid”, “whitegrid”, “dark”, “white”, “ticks”}) – Which seaborn style to apply to the plot? Default is whitegrid.
- sns_context ({“paper”, “notebook”, “talk”, “poster”}) – Which seaborn context to use? Controls the scaling of plot elements such as tick labels and the legend. Default is talk.
- despine (Bool) – Remove the top and right axes from the plot? Default is True.
- min_quantile (float (>0.0 and <1.0, default = 0.001)) – Clip data that is less than this quantile.
- max_quantile (float (>0.0 and <1.0, default = 1.00)) – Clip data that is greater than this quantile.
- lim (Dict(Str : (float, float))) – Set the range of each channel’s axis. If unspecified, assume that the limits are the minimum and maximum of the clipped data
- alpha (float (default = 0.02)) – The alpha blending value, between 0 (transparent) and 1 (opaque).
- axvlines_kwds (dict) – A dictionary of parameters to pass to ax.axvline
Notes
This uses a low-level API for speed; there are far fewer visual options that with other views.
- experiment (Experiment) – The
-